Supplementing vConTACT2 Clusters
vConTACT2, available here, is a fantastic tool for inferring the taxonomy of viral sequences. This pipeline clusters the user’s sequences with a RefSeq database based upon shared protein clusters.
As the RefSeq database contains relatively few genomes, more context can be given to a network by adding more reference genomes.
Phage genomes are collected each month and made publicly available here. Their genbank accessions are then used to extract useful information, which we (@CyanoNey) recently used to produce useful annotation files for phage phylogenetic trees, including viral family and bacterial host, available here.
Proteins were predicted on 12,892 phage genomes (30/05/2020) using Prodigal and input files for vConTACT2 were produced.
Simply combine these protein sequence fasta and gene-to-genome mapping files with your own, and they will be added to the network:
- Protein sequence file – available here
- Gene-to-genome mapping file – available here
(Warning: files are quite large)
Quickly Identifying Reference Genera
To quickly identify clusters of reference genomes at the genus, we (@RyanCookAMR) have produced a mapping file that will colour in these reference sequences:
- Annotation File – available here
To use the annotation file:
- Load your network into Cytoscape
- File -> Import -> Table from File…
- Then move to the “Style” portion of the control panel
- Fill Colour -> Mapping (middle box) -> Column = Colour_Hex -> Mapping = Passthrough Mapping
And the resultant network will look something like this:
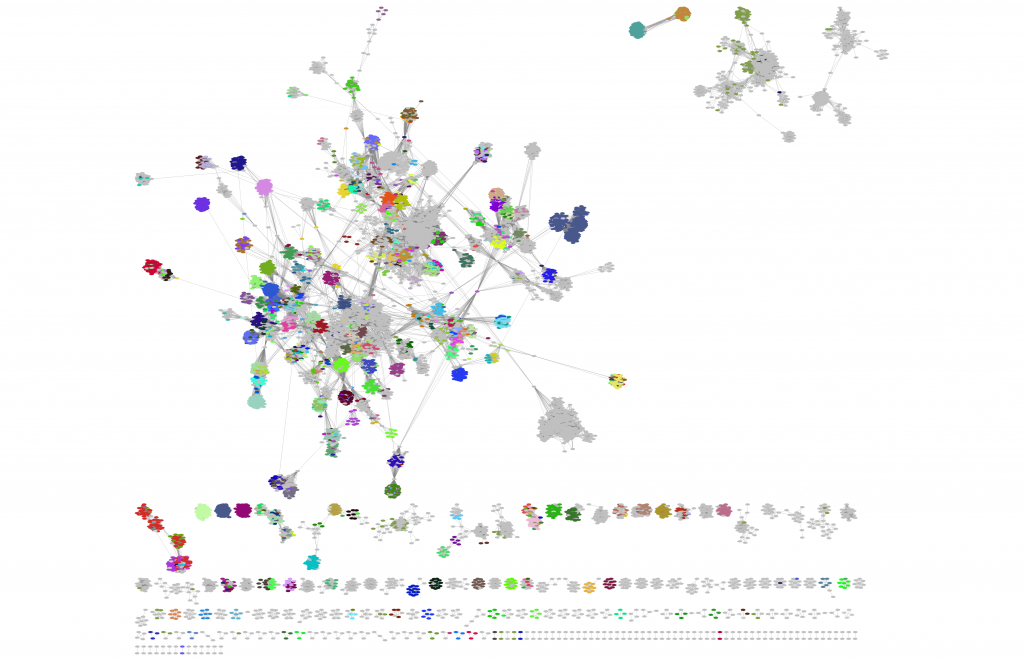
Alternatively, you can choose your own colours for specific genera of interest by selecting Column = Subfamily/Genera -> Mapping = Discrete Mapping, and then manually selecting colours.
Happy clustering!
