***As from October 2018 monthly updates will each have their own page***
This page will remain as it contains details of how genomes were extracted
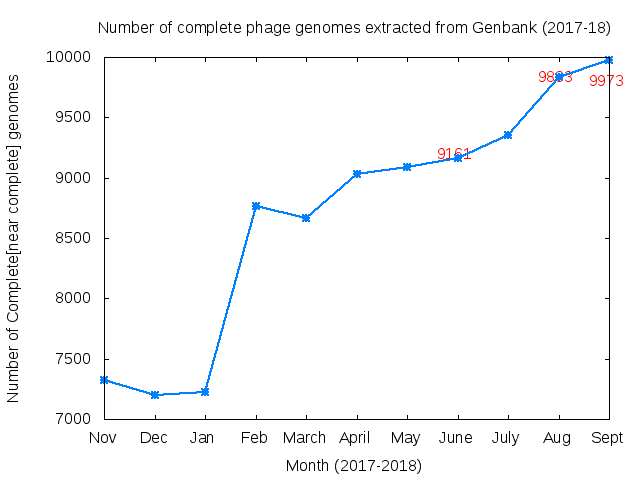
Table of >9000 Complete Bacteriophage genomes extracted from Genbank on 31st May 2018. The number of genomes has increased by only ~50 on last month. Based on feedback we have excluded a number of genomes from last month that were not complete but passed through our automated filtering system. The large increase this last month seems in part due to the submission of predicted prophages from bacterial genomes – but without experimental evidence they are functional.
Number of complete (near complete- see below) genomes .
We used ” esearch -db nucleotide -query \”gbdiv_PHG\”[prop] |efetch -format gb > phages.gb ” to first pull out all phage nucleotide data.
Then filtered this file for
“Complete” & “Genome|chromosome” or
“Sequence Length > 10000” & “Complete” & “Sequence|genome”
or Accession Number matches – a list of accessions we have manually curated of complete bacteriophage genomes that don`t match the above criteria. We updated these parsing parameters after helpful feedback that pointed out we were missing some phages. We think this allows the extraction of all complete (or near complete) bacteriophage genomes, whilst excluding fragments of genomes. We have manually looked at the list of descriptions we extract to identify obvious fragments eg 21 kb region of a cyanophage genome. With further rules to exclude these specific fragments.
Thanks to Kelly Williams for highlighting and then helping filter out some further issues. The table below will contain duplicates if the phage has been sequenced multiple times, in an experimental evolution experiment for instance. It also contains both a REFSEQ and the original Genbank entry file, so the number of unique genomes will be less. Using the phage description as an identifier this is 5446 genomes (from March).
Again thanks to Kelly Williams, who has found more complete phage genomes that are not identified by “gbdiv_PHG” in Genbank or are in the ENV database. The accessions for these genomes are
FN436269,NC_029032,NC_023585,NC_028693,NC_021793,NC_025471,NC_026606,NC_008355,NC_028651,NC_028998,NC_029103,NC_027984,NC_006938,HQ157199,NC_028990,NC_025444,HQ157198,KT626047,NC_028658,NC_014322,NC_028671,NC_024711,NC_028992,NC_029020,NC_029016,NC_029027,NC_029011
I will add these to our database next time we update it
Thanks to feedback from Andrew Kropinski we have removed to some further non-complete genomes.
Flat files are here for those wanting to batch download the entire list
- Complete_Bacteriophage_genomes_OCT_2017
- Complete Phage Genomes 12 Nov 2017
- Complete_phage_genomes_March2018
- 1April2018_phages.complete_genomes
- 31May2018_phages.complete_genomes_accession
- 30Jun2018_phages.complete_genomes_accession
Flat files with REFSEQ entries removed
Flat files with only REFSEQ entries – This is as extracted from this dataset for phages that have a RefSeq accession
[table “22” not found /]