Antibiotic resistance genes have been found in the viral fraction from a number of different environments including the murine gut (Modi et al. 2013), activated sludge (Parsley et al. 2010) and feaces from farm animals (Colomer-Lluch et al. 2011). Building on this previous research Slaweks PhD project will look at the role of bacteriophages as a reservoir of antibiotic resistance genes in the marine environment. This will involve metagenomics and culture bases studies developing Vibrio spp as a model phage-host system.
Images of some of the Vibrio spp isolated by Slawek and provided by collaborator Luigi Vezzulli
.
Progress
During his project Slawek has isolated a collection of phages that infected a broad range of Vibrio spp. These model systems are now being studied to further understand the role of vibriophages in horizontal gene transfer , in particular antibiotic resistance genes.
Prophage prediction
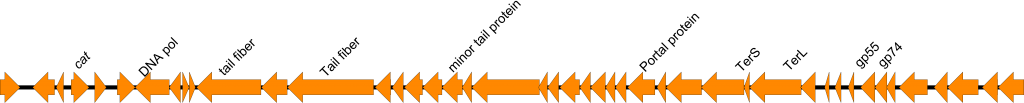
In addition to isolation of vibriophages, we have have been predicting the presence of prophages in Vibrio genomes. To date we have predictions for > 9000 vibrio genomes, with > 10000 prophage like elements. We have found the presence of antibiotic resistance genes on these prophages. An example is below of cat gene on a predicted prophage
References
Modi, S. R., Lee, H. H., Spina, C. S. & Collins, J. J. (2013) Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature, 499 (7457): 219-222.
Parsley, L. C., Consuegra, E. J., Kakirde, K. S., Land, A. M., Harper, W. F., Jr. & Liles, M. R. (2010) Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl Environ Microbiol, 76 (11): 3753-3757.
Colomer-Lluch, M., Imamovic, L., Jofre, J. & Muniesa, M. (2011) Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob Agents Chemother, 55 (10): 4908-4911
